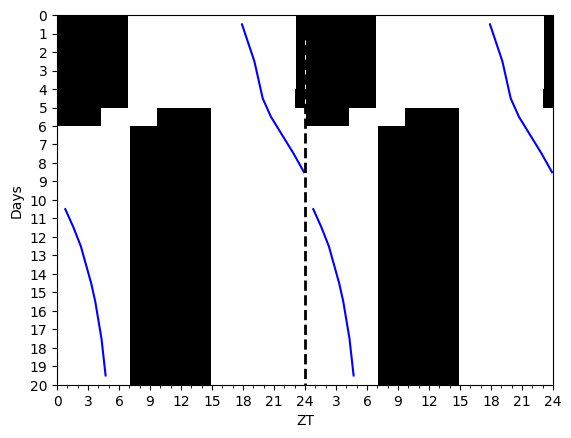
slam_shift = LightSchedule.SlamShift(shift=12.0, lux=500.0, before_days=2)
time = np.arange(0.0, 15*24.0, 0.10)
light_values = slam_shift(time)
# Run this for a range of period parameters
batch_dim = 50
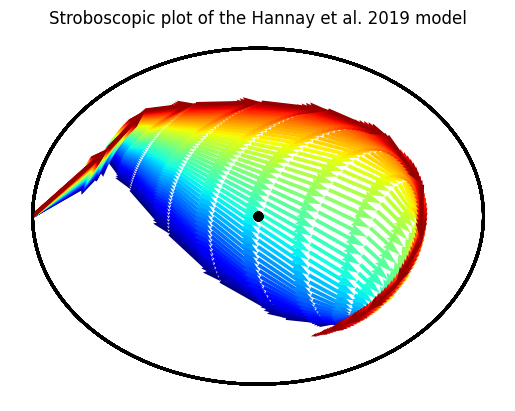
hmodel = Hannay19({'tau': np.linspace(23.5, 24.5, batch_dim)})
initial_state = np.array([1.0, np.pi, 0.0]) + np.zeros((batch_dim, 3))
initial_state = initial_state.T
trajectory = hmodel(time, initial_state, light_values)
ax = plt.gca()
cmap = plt.get_cmap('jet')
for idx in range(trajectory.batch_size):
Stroboscopic(ax,
time,
trajectory.states[:, 0, idx],
trajectory.states[:, 1, idx],
period=24.0,
lw=0.50,
color=cmap(idx/batch_dim));
plt.title("Stroboscopic plot of the Hannay et al. 2019 model");